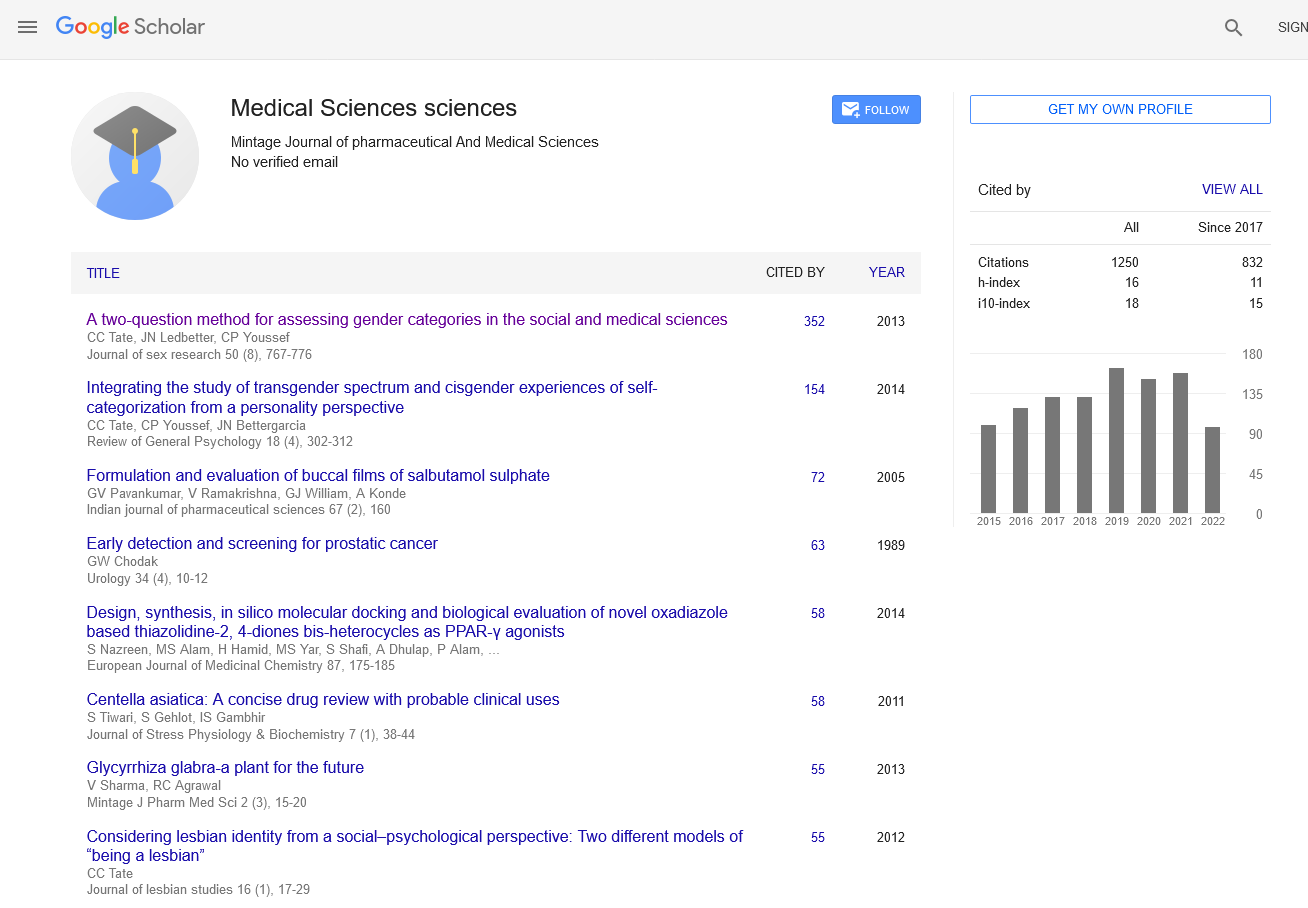
THE GENETIC VARIATION IN ANCESTRAL POPULATIONS AND SPECIES
Short Communication - (2023) Volume 12, Issue 1
Introduction
Since the beginning of the genome period, established researchers has depended on a solitary ‘reference’ genome for every species, which is utilized as the reason for a large number of hereditary examinations, including investigations of variety inside and across species. As sequencing costs have dropped, a huge number of new genomes have been sequenced, and researchers have come to understand that a solitary reference genome is deficient for some reasons.
Description
Specifically, by examining a different arrangement of people, one can start to gather a container genome: an assortment of all the DNA successions that happen in animal groups. Here we survey endeavors to make container genomes for a scope of animal varieties, from microorganisms to people, and we further consider the computational strategies that have been proposed to catch, decipher and look at skillet genome information. As researchers proceed to study and index the genomic variety across human populaces and start to gather a human skillet genome, these endeavors will expand our ability to interface variety to human variety, sickness and then some [1].
Along with cutting edge sequencing strategies, progresses in metagenomics and single-cell genomics have lifted this restriction, accommodating an undeniably fair portrayal of the worldwide prokaryote variety. Propels in computational genomics followed the advancement of genome sequencing, regardless of whether sometimes lingering behind. A few significant new parts of microorganisms and archaea were found, including Asgard archaea, the evident nearest family members of eukaryotes and broad gatherings of microscopic organisms and archaea with little genomes remembered to be symbionts of different prokaryotes. Near investigation of various prokaryote genomes crossing a large number of transformative distances changed the calculated underpinnings of microbial science, replacing the idea of species genomes with fixed quality sets with that of dynamic pangenomes and the thought of a solitary Tree of Life (ToL) with a measurable tree-like pattern among individual quality trees. Steps were likewise made towards a hypothesis and quantitative laws of prokaryote genome development [2,3].
Borrowing both from populace hereditary qualities and phylogenetics, the field of populace genomics arose as full genomes of a few firmly related animal categories were accessible. Giving we can appropriately demonstrate arrangement advancement inside populaces going through speciation occasions, this asset empowers us to appraise key populace hereditary qualities boundaries, for example, familial populace sizes and split times. Moreover we can upgrade how we might interpret the recombination interaction and examine different particular powers. With the approach of re-sequencing advances, vast examples of variety in surviving populaces have now come to supplement this image, offering a rising ability to concentrate on later hereditary history.We examine the essential models of genomes in populaces, including speciation models for firmly related species. A significant point in our conversation is that a couple of complete genomes contain a lot of data about the entire populace [4].
Conclusion
The explanation being that recombination unlinks genomic locales, and hence a couple of genomes contain many sections with unmistakable chronicles. The test of populace genomics is to decipher this mosaic of accounts to deduce situations of demography and choice. We overview demonstrating systems for grasping hereditary variety in familial populaces and species. The hidden models expand on the coalescent with recombination process and acquaint further suppositions with scale the investigations to genomic informational collections.
Acknowledgement
The authors are very thankful and honoured to publish this article in the respective Journal and are also very great full to the reviewers for their positive response to this article publication.
Conflict of Interest
We have no conflict of interests to disclose and the manuscript has been read and approved by all named authors.
References
- Taylor MRG. Pharmacogegenetics of human beta-adrenergic receptors. Pharmacogenomics J 2007; 7:29-37.
- Herrington DM. Role of estrogen receptor-a in pharmacogenetics of estrogen action. Curr Opin Lipidol 2003; 14:145-150.
- Johnson JA, Lima JJ. Drug receptor/effector polymorphisms and pharmacogenetics: Current status and challenges. Pharmacogenetics 2003; 13:525-534.
- Zavratnik A, Prezelj J, Kocijancic A, et al. Exonic, but not intronic polymorphism of ESR1 gene might influence the hypolipemic effect of raloxifene. J Steroid Biochem Mol Biol 2006; 104:22-26.
Author Info
Vicente Quiny*Received: 30-Jan-2023, Manuscript No. mjpms-23-92851; , Pre QC No. mjpms-23-92851 (PQ); Editor assigned: 01-Feb-2023, Pre QC No. mjpms-23-92851 (PQ); Reviewed: 15-Feb-2023, QC No. mjpms-23-92851; Revised: 20-Feb-2023, Manuscript No. mjpms-23-92851 (R); Published: 27-Feb-2023, DOI: 10.4303/mjpms/236035
Copyright: This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

ISSN: 2320-3315
ICV :81.58